Analytics: Imaging
SLIM-Based High-Resolution Ion Mobility to Interrogate Glycopeptide Heterogeneity
What improvements could High Resolution Ion Mobility (HRIM) bring to separation ability, throughput and reproducibility, and researchers’ understanding of glycoprotein structures?
Amber Jones, Roxana McCloskey, and James Atwood at MOBILion Systems, Inc
A protein’s glycoforms dictate functional properties, such as half-life/clearance and downstream effect, or functions/binding partners. Given that most biologics are glycoproteins, researchers need to define the glycans attached and the site-specific microheterogeneity on these glycoproteins. Separating glycopeptides via chromatography is highly challenging, due to the high level of glycan heterogeneity, and often requires gradients that are extended over hours, frequently ending in coelution and an inability to assign unambiguous identification.
Recent advances in glycoscience have shone new light on glycans and their role as key metabolic, structural, and physical components in biological structures. Although less studied than other major building blocks of life – i.e., proteins, lipids, and nucleic acids – complex and diverse glycans are essential to life and are omnipresent in organisms from archaea to humans (1). Glycan analysis, or glycomics in which scientists elucidate the specific glycosylation patterns of biomolecules, has become an essential aspect of biomedical research, drug development, and biopharma quality control.
Glycan structures are inherently complex and heterogeneous. Though most mammalian glycans are built from around nine monosaccharide units, these monomers can be attached to one another through many different glycosidic linkages, including in a branched fashion (2). Collectively, the number of possible glycan structures quickly expands with each additional monomer and branching. It is estimated that for a hexasaccharide there are, in total, around 1,012 possible glycan structures, though they may not all occur in nature (3).
Much of the glycome are isomers and, therefore, it is vital that each structure is identified appropriately. Released glycan workflows are routinely employed for biopharmaceutical glycan analysis, as they provide details around the glycan composition that can impact efficacy and safety of drug products. However, glycopeptide workflows are emerging as an attractive alternative to released glycan analysis due to their lower cost and higher throughput.
The chosen technique for released glycan analysis has long been mass spectrometry (MS), as it offers the resolving power and sensitivity needed to elucidate specific glycan structures and glycoforms. However, to achieve meaningful resolution of glycan isomers, separation techniques have been employed ahead of MS analysis.
Traditional liquid chromatography (LC) coupled with MS-based methods for determining glycan structures are time-consuming, complex, and often not effective enough to address the composition and linkage diversity of glycans adequately. Ion mobility has emerged as a powerful tool to resolve glycan and glycopeptide isomers and to provide unambiguous identifications.
Current instruments might provide sufficient throughput, resolution, or ease-of-use, but never all three at the same time. On a practical level, LC separation of highly similar biomolecules – like many glycans are – often requires extended run times, creating a potential bottleneck in analytical workflows. Even with long run times, a high percentage of glycopeptides coelute, which complicates or even prevents full structural assignment (4).
So, while LC-MS remains the standard workflow for glycopeptide analysis, there is a pressing need for techniques with better resolving power and faster turnaround.
HRIM-MS
High Resolution Ion Mobility-Mass Spectrometry (HRIM-MS) based on Structures for Lossless Ion Manipulation (SLIM) has emerged as a promising separation strategy in glycopeptide analysis (5). With HRIM, as with other ion mobility technologies, ionised molecules are separated in the gas phase, based on physical characteristics such as size and shape.
Unique to SLIM-based HRIM-MS is the use of printed circuit boards (PCBs) (Figure 1) to create an exceptionally long separation path within a small footprint. SLIM has the ability to make ions turn around corners instead of traveling on a straight path, yielding a 13m single-pass flight path. The electric fields that propel the ions along the separation path also prevent them from striking surfaces while moving, preventing any losses along their way. This allows them to be separated with very high resolution and in a timeframe of minutes, without loss of sensitivity (6). With the straightforward set up of SLIM-based HRIM, there is no need to optimise columns, flow rates, or liquid components as with LC. In addition, SLIM-based HRIM achieves rapid separations with data collection in the order of milliseconds to seconds.
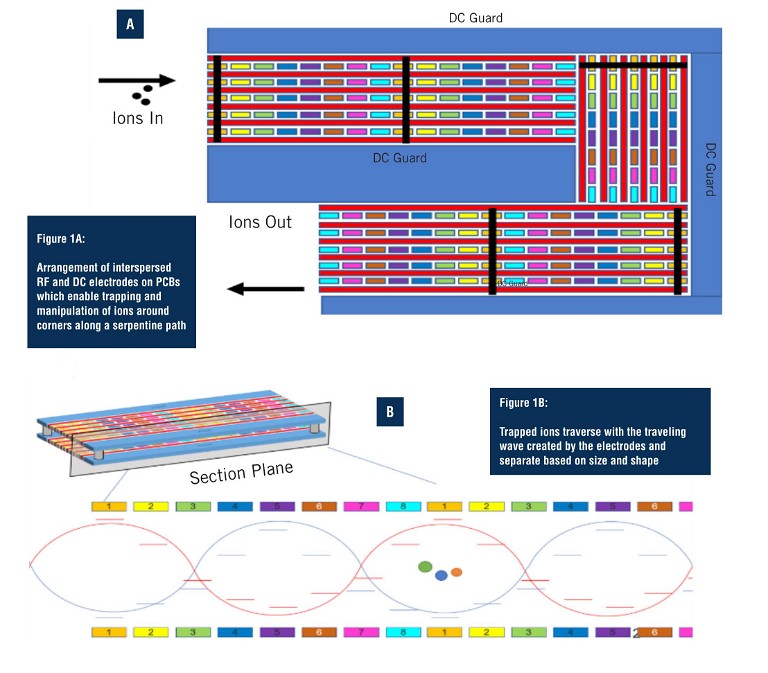
Much has been written about how the most advanced HRIM approaches boost resolving power by expanding the pathlength (7). When combined with MS, the technique offers tremendous potential for seamless separation of released glycans and glycopeptides with composition and structural determination to support biopharmaceutical characterisation, biomedical, and clinical research (8, 9). HRIM allows researchers to monitor released glycan and glycopeptide structural diversity with added specificity, higher reproducibility, and better resolution than current workflows. The HRIM-MS platform was employed to investigate SLIM-based HRIM-MS for improved separation ability and throughput for glycopeptide workflows.
Method and Results
The glycoprotein NISTmAb was reduced, carboxyamidomethyled, and enzymatically digested with trypsin. The resulting sample was analysed via reverse phase, high performance liquid chromatography (HPLC)-HRIM-MS, to resolve as many glycopeptide species as possible. The analysis was performed on the HRIM platform (MOBIE, MOBILion Systems) coupled to a 6545XT Q-ToF platform (Agilent, Santa Clara, CA, USA). Glycopeptide ID was performed on a HRIM Peptide Workflow module (Protein Metrics Inc., Cupertino, CA, USA).
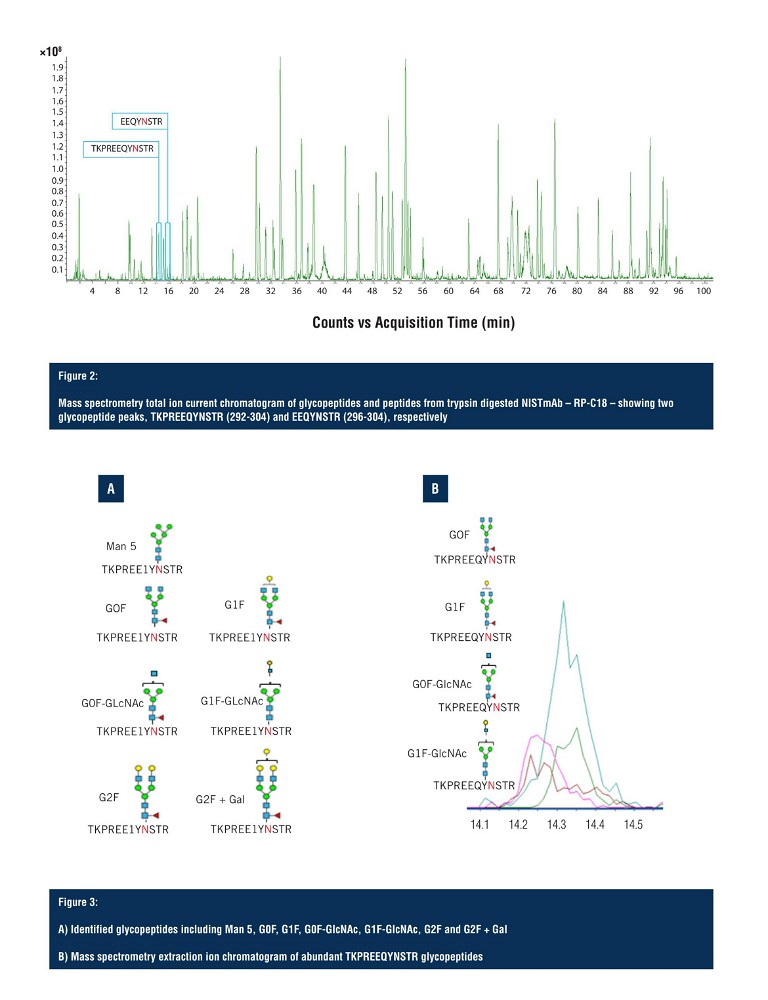
In Figure 2, the total ion chromatogram demonstrates adequate separation of the trypsin digested NISTmAb peptides, and indicates the region of elution for the two glycopeptide species. The first glycopeptide peak to arrive is KPREEQYNSTR (292-304), along with all its glycoforms, and the second is EEQYNSTR (296-304), along with all its glycoforms. However, as shown on the total ion current chromatogram, the glycopeptides elute in a very small window, making detection and resolution of multiple glycopeptide glycoforms challenging.
Both glycopeptide species elute between 14 and 16 minutes with the first glycopeptide, KPREEQYNSTR, being observed with seven glycoforms, identified via LC-MS (Figure 3A) with four of those being assigned as ambiguous compositions (Figure 3B). All seven glycoforms eluted within a 30 second window, with limited resolution in the LC domain, making unambiguous structural assignment challenging.
The HRIM platform is capable of performing a full m/z range HRIM separation per second, providing multiple ion mobility scans across the 30 second elution window, making it particularly amenable to LC-MS-based analyses and glycopeptide workflows. The amount of data acquired is greater and often more impactful compared to the same period using LC alone.
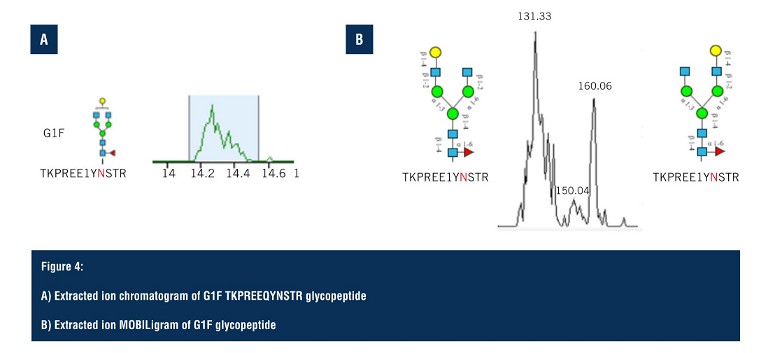
Using SLIM-based HRIM-MS for glycopeptide analysis workflows allows the time-to-result to be dramatically shortened, all whilst improving characterisation of isomeric mixtures. Pulling out the G1F glycoform isomer (Figure 4A), the extracted ion chromatogram predominantly shows a single peak but looking closely, there are, in fact, two separate species that are hard to distinguish due to glycan heterogeneity. On the extraction ion mobiligram (Figure 4B), the distribution shown mirrors that of the G1F released glycan HRIM profile with a bimodal distribution.
The earlier arriving species is assigned to the galactose attached to the alpha 1,3 branch, and the later arriving species is assigned to the glycopeptide contain the galactoseattached to the alpha 1,6 branch. Glycopeptide separations are achieved via changes in gas phase size as a result of either positional or linkage differences on the glycan moiety, enabling glycopeptide isomers to be separated that are often unresolved via LC alone. From the HRIM generated extracted ion mobiligrams, extracted ion chromatograms can be generated for specific regions of the mobiligram to monitor LC retention time profiles as a function of mobility arrival time. This is particularly helpful in assigning glycopeptide isomer identifications.
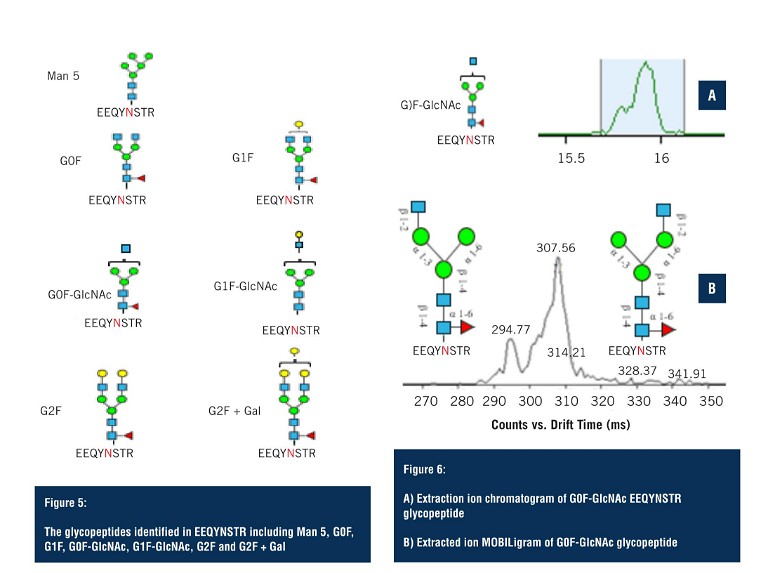
The second group of glycopeptides were identified on the fully tryptic peptide EEQYNSTR. Although in lower abundance, LC-MS was still able to detect seven glycoforms (Figure 5), with multiple ambiguous identifications.
Applying the analysis as performed on the G1F glycopeptide – the extracted ion chromatogram of the G0F glycopeptide – again shows a major peak and, upon closer inspection, two species can be observed that are unresolved in the LC domain (Figure 6A).
By applying HRIM, the extracted ion mobiligram reveals a bimodal distribution due to glycan heterogeneity at the addition site of the terminal GlcNAc. Changes in the size of the glycopeptide as a result of these positional isomers allows them to be separated via HRIM, and assists in making structural assignments that would not otherwise have been possible with LC-MS alone.
Conclusion
Using ion mobility separations, the length of the ion separation path is crucial to achieving the high degree of separation required to gain adequate resolution of glycans and glycopeptides with challenging structural diversity.
The results discussed above indicate that separation using the SLIM-based HRIM-MS platform enables separation of isomeric glycopeptide mixtures differing only by the structure of the glycan. This provides more structural information than conventional LC-based separation workflows, in much less time.
“ Current instruments might provide sufficient throughput, resolution, or ease-of-use, but never all three at the same time ”
Compared to other ion mobility separation platforms, SLIM-based HRIM-MS provides the highest resolution single-pass separation across the broadest m/z range in the least amount of time. Translating this into real life, this capability enables more specificity in glycoprotein analysis, a critical class of molecules for biotherapeutics development and discovery research.
References
1. Varki A, Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells, Cold Spring Harb Perspect Biol 3(6), 2011; doi: 10.1101/ cshperspect.a005462.
2. Seeberger PH, Monosaccharide diversity, 2017. In: Varki A et al, Essentials of Glycobiology, Chapter 2, 3rd edition, 2015-2017; doi: 10.1101/glycobiology.3e.002
3. Laine RA, A calculation of all possible oligosaccharide isomers both branched and linear yields 1.05 x 1012 structures for a reducing hexasaccharide: The Isomer Barrier to development of single-method saccharide sequencing or synthesis systems, Glycobiology 4(6): pp759-767, 1994; doi:10.1093/ glycob/4.6.759
4. Rudd P et al, Glycomics and Glycoproteomics, 2017. In: Varki A et al, Essentials of Glycobiology, Chapter 51, 3rd edition, 2015-2017, https://www.ncbi.nlm.nih.gov/books/NBK453015/ doi: 10.1101/glycobiology.3e.051
5. Hofmann J et al, Glycan Analysis by Ion Mobility-Mass Spectrometry, Angew Chem Int Edit 56(29): pp8342-9, 2017; doi: 10.1002/anie.201701309.
6. L Deng et al, Ultra-high resolution ion mobility separations utilizing traveling waves in a 13 m serpentine path length structures for lossless ion manipulations module, Anal Chem (88): pp8957–8964, 2016
7. Hollerbach AL et al, Ultra-High-Resolution Ion Mobility Separations Over Extended Path Lengths and Mobility Ranges Achieved using a Multilevel Structures for Lossless Ion Manipulations Module, Anal Chem 92(11): pp7972–9, 2020
8. Arnaud CH, Resolving power for the people: Ion mobility-mass spec expands its offerings, C&EN 98(20), 2020
9. Wormwood KL et al, The potential for Ion Mobility in pharmaceutical and clinical analyses, Advancements of Mass Spectrometry in Biomedical Research: pp299-316, 2019

A graduate of Virginia Commonwealth University, US, Amber Jones has spent the past 15 years in the life science market supporting clinical and commercial manufacturing of biotherapeutics.
With a focus on cell and gene therapy for the past decade, Amber has written publications and aided in regulatory filings to help bring lifesaving therapies to market with various global partners.
Recently Amber has turned to analytics and characterisation of complex samples. As product manager at MOBILion Systems Inc, Amber aims to introduce SLIM technology to those who need the unprecedented resolution and characterisation capabilities it provides.

After graduating from Simmons University in Boston, US, Roxana McCloskey spent several years in genomics research at Brigham and Women’s Hospital, before moving in to sales and marketing in the sample prep and mass spec markets.
As Marketing Communications Manager at MOBILion, Roxana is focused on introducing MOBIE and how HRIM-MS is addressing characterisation gaps and providing enabling solutions to the mass spec community.

James Atwood obtained his PhD in Analytical Chemistry, with a focus on high throughput bottom-up proteomics and glycomics, from the University of Georgia, US.
He brings over 15 years of experience leading businesses that develop and supply products and services for the life science markets. He has a passion for scientific product commercialisation with a particular interest in proteomics and glycomics. As MOBILion’s SVP of Biopharma and Omics, James is focused on bringing MOBILion technology into the ‘multiomics’ space.